Minimum Abundance Threshold
With less samples multiplexed per sample than our own work (which guided the default settings), these samples were sequenced at much higher depth:
$ cut -f 1,2,5 metadata.tsv
<SEE TABLE BELOW>
As a table:
run_accession |
library_name |
read_count |
|---|---|---|
SRR7109326 |
m6-stds |
817764 |
SRR7109327 |
m6-301-1 |
890561 |
SRR7109328 |
m6-mock3-32000b |
943839 |
SRR7109329 |
m6-766-1 |
840068 |
SRR7109330 |
m6-744-2 |
704173 |
SRR7109331 |
m6-500-1 |
911793 |
SRR7109341 |
m6-712-2 |
872265 |
SRR7109342 |
m6-500-2 |
879762 |
SRR7109343 |
m6-757-1 |
903886 |
SRR7109344 |
m6-757-2 |
1210627 |
SRR7109345 |
m6-mock3-16000 |
922440 |
SRR7109406 |
m6-712-1 |
897159 |
SRR7109408 |
m6-744-1 |
778090 |
SRR7109409 |
m6-mock3-32000a |
1125275 |
SRR7109411 |
m6-766-2 |
785776 |
SRR7109412 |
m6-755-1 |
957067 |
SRR7109414 |
m6-736-1 |
998817 |
SRR7109415 |
m6-301-2 |
1181567 |
SRR7109417 |
m6-755-2 |
1071829 |
SRR7109418 |
m6-736-2 |
919363 |
SRR7109420 |
m6-SynMock |
1299238 |
The defaults are an absolute abundance threshold of 100, and a fractional
threshold of 0.1% (i.e. -a 100 -f 0.001). After merging overlapping reads
and primer matching we could expect over 650,000 reads per m6 sample, giving a
threshold over 650 reads.
So, with this coverage the default fractional abundance threshold of 0.1%
(i.e. -f 0.001) makes the default absolute abundance threshold of 100
(i.e. -a 100) redundant. However, on this dataset our defaults are quite
cautious, and control samples can help set thresholds objectively.
In this dataset there is a single synthetic control for m6 sequencing run,
library SynMock aka SRR7109420. We can tell THAPBI PICT at the command
line to use this to set the fractional abundance threshold via -y or
--synctrls, and/or set the absolute abundance threshold via -n or
--negctrls (with a list of control file names). It turns out however that
with the default thresholds the control is clean (no unwanted non-synthetic
ITS2 reads).
Using the defaults
The first step in run.sh is to run the pipeline with the default abundance
thresholds (stricter than the alternatives analyses below), giving just a few
hundred unique ITS2 sequences:
$ grep -c "^ITS2" summary/defaults.ITS2.tally.tsv
360
$ grep -c "^ITS2" summary/defaults.ITS2.reads.1s5g.tsv
360
Look at summary/defaults.ITS2.samples.1s5g.xlsx or working at the command
line with the TSV file:
$ cut -f 1,7,9,11-12,14-15 summary/defaults.ITS2.samples.1s5g.tsv
<SEE TABLE BELOW>
As a table:
#sample_alias |
Cutadapt |
Threshold |
Max non-spike |
Max spike-in |
Accepted |
Unique |
|---|---|---|---|---|---|---|
301-1 |
807956 |
808 |
348111 |
0 |
528957 |
38 |
301-2 |
1108129 |
1109 |
457440 |
0 |
778850 |
31 |
500-1 |
819468 |
820 |
289229 |
0 |
516474 |
30 |
500-2 |
813470 |
814 |
214155 |
0 |
529967 |
34 |
712-1 |
820146 |
821 |
131937 |
0 |
533310 |
56 |
712-2 |
796363 |
797 |
299240 |
0 |
520290 |
34 |
736-1 |
943427 |
944 |
349965 |
0 |
669563 |
36 |
736-2 |
854919 |
855 |
282132 |
0 |
609025 |
25 |
744-1 |
706659 |
707 |
358089 |
0 |
493209 |
20 |
744-2 |
651528 |
652 |
136471 |
0 |
452421 |
36 |
755-1 |
887650 |
888 |
462493 |
0 |
616322 |
27 |
755-2 |
982087 |
983 |
589120 |
0 |
669602 |
17 |
757-1 |
835431 |
836 |
281533 |
0 |
578198 |
35 |
757-2 |
1099959 |
1100 |
224635 |
0 |
742540 |
28 |
766-1 |
792260 |
793 |
526535 |
0 |
583643 |
16 |
766-2 |
711176 |
712 |
251097 |
0 |
469397 |
26 |
BioMock |
866253 |
867 |
56120 |
0 |
591947 |
23 |
BioMock |
846519 |
847 |
65686 |
0 |
585715 |
23 |
BioMock |
1023231 |
1024 |
84748 |
0 |
698170 |
22 |
BioMockStds |
736334 |
737 |
35300 |
0 |
521693 |
26 |
SynMock |
1199806 |
1200 |
0 |
103014 |
862950 |
18 |
The SynMock control is clean, no non-spike-in reads passed the default
abundance thresholds.
So, there is scope to lower the default thresholds - but how low? We will start by reproducing the Illumina part of Figure 6, which was based on the m6 MiSeq sequencing run. This figure explores tag-switching in the demultiplexing, and in the authors’ analysis goes as low as 5 reads.
Excluding only singletons
The run.sh example continues by running the pipeline on the m6 dataset with
-f 0 -a 2 to accept everything except singletons (sequences which are only
seen once in a sample; including them gives about ten times as many unique
sequences which slows everything down). Also, this analysis does not use the
synthetic control to raise the threshold on the rest of the samples - we want
to see any low level mixing. We then can compare our sample report against
Figure 6.
Looking at the unique reads in the FASTA file, tally table, or in the reads report with metadata, we have nearly 200 thousand ITS2 sequences:
$ grep -c "^ITS2" summary/a2.ITS2.tally.tsv
196480
$ grep -c "^ITS2" summary/a2.ITS2.reads.onebp.tsv
196480
Look at summary/a2.ITS2.samples.onebp.xlsx or working at the command line
with the TSV file:
$ cut -f 1,5-7,11-12,14-15 summary/a2.ITS2.samples.onebp.tsv
<SEE TABLE BELOW>
As a table:
#sample_alias |
Raw FASTQ |
Flash |
Cutadapt |
Max non-spike |
Max spike-in |
Accepted |
Unique |
|---|---|---|---|---|---|---|---|
301-1 |
890561 |
812674 |
807956 |
348111 |
0 |
687950 |
12638 |
301-2 |
1181567 |
1113606 |
1108129 |
457440 |
0 |
977003 |
13319 |
500-1 |
911793 |
823392 |
819468 |
289229 |
0 |
689174 |
14249 |
500-2 |
879762 |
817277 |
813470 |
214155 |
0 |
699634 |
12851 |
712-1 |
897159 |
823034 |
820146 |
131937 |
0 |
703189 |
17574 |
712-2 |
872265 |
800475 |
796363 |
299240 |
0 |
683057 |
13937 |
736-1 |
998817 |
948348 |
943427 |
349965 |
15 |
834461 |
12993 |
736-2 |
919363 |
858915 |
854919 |
282132 |
0 |
757097 |
9625 |
744-1 |
778090 |
710762 |
706659 |
358089 |
0 |
614988 |
7936 |
744-2 |
704173 |
654661 |
651528 |
136471 |
0 |
564238 |
8650 |
755-1 |
957067 |
891942 |
887650 |
462493 |
15 |
782052 |
12142 |
755-2 |
1071829 |
987280 |
982087 |
589120 |
0 |
848793 |
10587 |
757-1 |
903886 |
839105 |
835431 |
281533 |
0 |
725057 |
12729 |
757-2 |
1210627 |
1105530 |
1099959 |
224635 |
0 |
950457 |
15819 |
766-1 |
840068 |
794475 |
792260 |
526535 |
0 |
712126 |
7519 |
766-2 |
785776 |
714894 |
711176 |
251097 |
0 |
606887 |
11189 |
BioMock |
943839 |
872263 |
866253 |
56120 |
0 |
744007 |
17274 |
BioMock |
922440 |
859262 |
846519 |
65686 |
0 |
733784 |
16676 |
BioMock |
1125275 |
1047383 |
1023231 |
84748 |
3 |
884514 |
18416 |
BioMockStds |
817764 |
740627 |
736334 |
35300 |
0 |
628576 |
17202 |
SynMock |
1299238 |
1204532 |
1199806 |
187 |
103014 |
1043525 |
14234 |
Here SynMock (SRR7109420) is the synthetic control, and it has some
non-spike-in reads present, the most abundant at 187 copies. Conversely,
samples 755-1 (SRR7109412), 736-1 (SRR7109414), and one of the
BioMock samples (SRR7109409) have trace levels of unwanted synthetic
spike-in reads, the most abundant at 15, 15 and 3 copies respectively. The
counts differ, but these are all samples highlighted in Figure 6 (sharing the
same Illumina i7 or i5 index for multiplexing). We don’t see this in the other
BioMock samples, but our pipeline appears slightly more stringent.
As percentages, 187/1199806 gives 0.0156% which is nearly ten times lower than our default of 0.1%. The numbers the other way round are all even lower, 15/462496 gives 0.003%, 15/349965 gives 0.004%, and 3/1023234 gives 0.003%.
Using the synthetic control
Next the run.sh example uses the SynMock synthetic control to
automatically raise the fractional abundance threshold from zero to 0.015% by
including -a 100 -f 0 -y raw_data/SRR7109420_*.fastq.gz in the command line.
This brings down the unique sequence count enough to just over three thousand,
allowing use of a slower but more lenient classifier as well:
$ grep -c "^ITS2" summary/ctrl.ITS2.tally.tsv
3097
$ grep -c "^ITS2" summary/ctrl.ITS2.reads.1s5g.tsv
3097
Look at summary/ctrl.ITS2.samples.1s5g.xlsx or working at the command line
with the TSV file:
$ cut -f 1,7,9,11-12,14-15 summary/ctrl.ITS2.samples.1s5g.tsv
<SEE TABLE BELOW>
Note we now get a threshold column showing the absolute threshold applied to each sample (using the inferred percentage), all above the absolute default of 100. You can see the total accepted read count has dropped, and the number of unique sequences accepted has dropped even more dramatically:
#sample_alias |
Cutadapt |
Threshold |
Max non-spike |
Max spike-in |
Accepted |
Unique |
|---|---|---|---|---|---|---|
301-1 |
807956 |
126 |
348111 |
0 |
579502 |
262 |
301-2 |
1108129 |
173 |
457440 |
0 |
829870 |
189 |
500-1 |
819468 |
128 |
289229 |
0 |
568336 |
228 |
500-2 |
813470 |
127 |
214155 |
0 |
578432 |
215 |
712-1 |
820146 |
128 |
131937 |
0 |
569100 |
181 |
712-2 |
796363 |
125 |
299240 |
0 |
570488 |
243 |
736-1 |
943427 |
148 |
349965 |
0 |
708900 |
183 |
736-2 |
854919 |
134 |
282132 |
0 |
653753 |
220 |
744-1 |
706659 |
111 |
358089 |
0 |
540597 |
273 |
744-2 |
651528 |
102 |
136471 |
0 |
472785 |
129 |
755-1 |
887650 |
139 |
462493 |
0 |
694273 |
340 |
755-2 |
982087 |
154 |
589120 |
0 |
754928 |
338 |
757-1 |
835431 |
131 |
281533 |
0 |
610579 |
171 |
757-2 |
1099959 |
172 |
224635 |
0 |
781212 |
142 |
766-1 |
792260 |
124 |
526535 |
0 |
648524 |
301 |
766-2 |
711176 |
111 |
251097 |
0 |
508838 |
205 |
BioMock |
866253 |
136 |
56120 |
0 |
607401 |
77 |
BioMock |
846519 |
132 |
65686 |
0 |
603186 |
82 |
BioMock |
1023231 |
160 |
84748 |
0 |
718660 |
85 |
BioMockStds |
736334 |
115 |
35300 |
0 |
526317 |
48 |
SynMock |
1199806 |
100 |
187 |
103014 |
885051 |
113 |
Note that Palmer et al. (2018) apply a threshold to individual sequences, but the thresholding strategy in THAPBI PICT applies the fractional threshold to all the samples (given in the same sub-folder as input, so you can separate your MiSeq runs, or your PCR plates, or just apply a global threshold).
In fact, looking at the read report summary/ctrl.ITS2.reads.1s5g.tsv it is
clear that while this threshold may have excluded Illumina tag-switching, it
has not excluded PCR noise - there are hundreds of low abundance sequences
unique to a single sample. To address that we would have to use a considerably
higher threshold, and the default 0.1% is a reasonable choice here, or apply a
denoising algorithm like UNOISE.
Threshold selection
Excluding only singletons is too lenient, but how does the the synthetic control inferred threshold (0.0156%) compare to the default (0.1%)?
Here are the classifier assessment values using the lower inferred threshold which allows a lot of PCR noise:
$ head -n 2 summary/ctrl.ITS2.assess.1s5g.tsv
<SEE TABLE BELOW>
As a table:
#Species |
TP |
FP |
FN |
TN |
sensitivity |
specificity |
precision |
F1 |
Hamming-loss |
Ad-hoc-loss |
|---|---|---|---|---|---|---|---|---|---|---|
OVERALL |
102 |
11 |
1 |
186 |
0.99 |
0.94 |
0.90 |
0.94 |
0.0400 |
0.105 |
Versus the stricter higher default abundance fraction which excludes most of the PCR noise:
$ head -n 2 summary/defaults.ITS2.assess.1s5g.tsv
<SEE TABLE BELOW>
As a table:
#Species |
TP |
FP |
FN |
TN |
sensitivity |
specificity |
precision |
F1 |
Hamming-loss |
Ad-hoc-loss |
|---|---|---|---|---|---|---|---|---|---|---|
OVERALL |
92 |
8 |
11 |
189 |
0.89 |
0.96 |
0.92 |
0.91 |
0.0633 |
0.171 |
You could use the assessment metrics to help decide on your preferred threshold, depending on the best tradeoff for your use-case.
Personally, of the these two I would pick the higher default threshold since it appears to exclude lots of PCR noise as seen in the edit graphs. With the default 0.1% threshold:
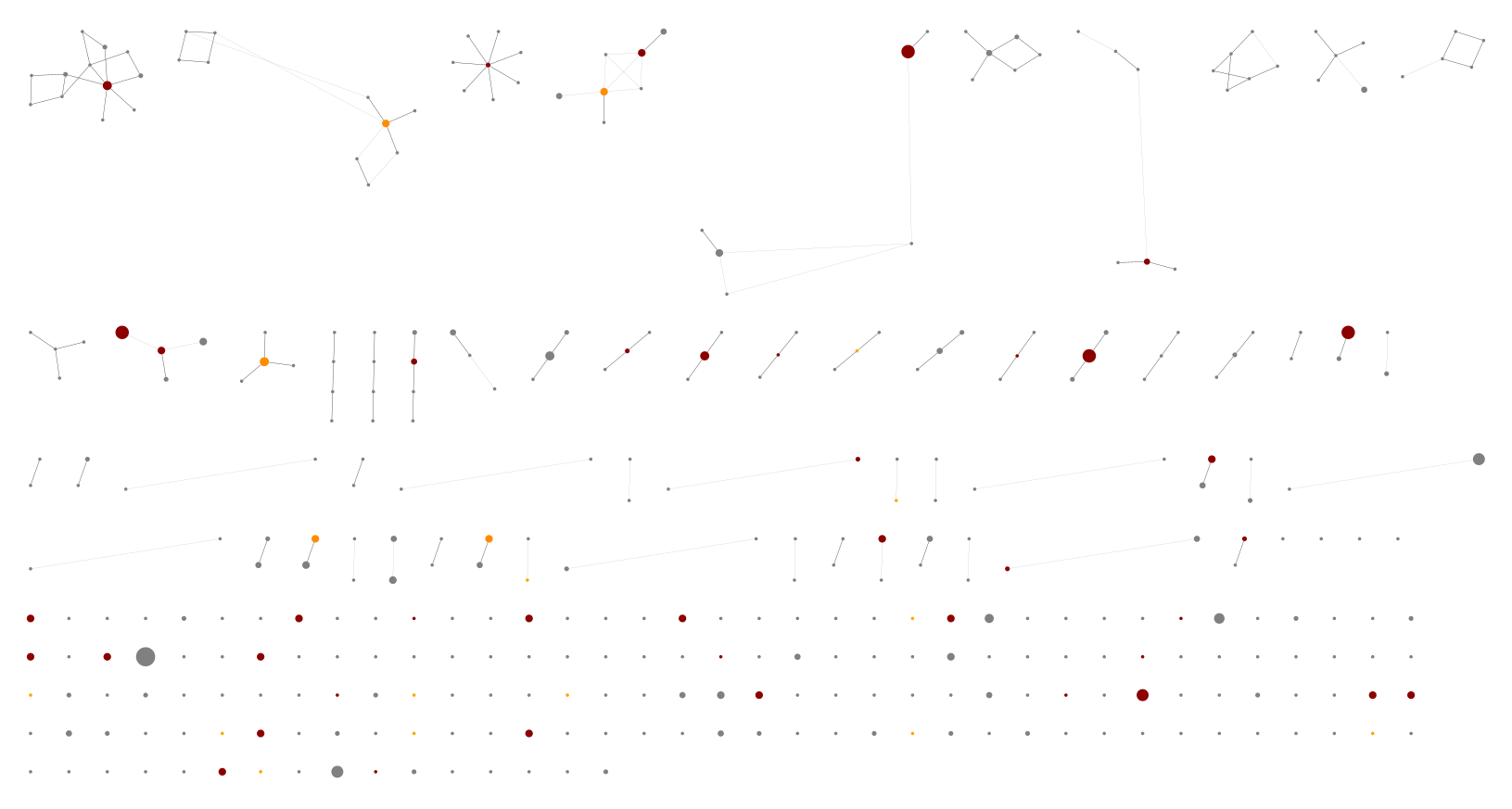
Using the lower threshold there are roughly ten times as many ASVs. The more common ASV nodes become the centre of a halo of 1bp variants, typically each seen in a single sample, which we attribute to PCR noise:

The best choice of threshold may lie somewhere in between?
Read-correction for denoising
Read-correction is an alternative or supplement to a stringent abundance filter
for removing the noise of sequence variants presumed to be PCR artefacts. Use
--denoise as part of the pipeline or sample-tally commands to enable our
implementation of the UNOISE algorithm (Edgar 2016).
Adding this to the control-driven abundance threshold example drops the total unique read count from over 3 thousand to just over 700:
$ grep -c "^ITS2" summary/ctrl_denoise.ITS2.tally.tsv
704
$ grep -c "^ITS2" summary/ctrl_denoise.ITS2.reads.1s5g.tsv
704
This gives an edit graph visually somewhere in between the examples above, with the obvious variant halos collapsed, but some of the more complex chains of variants still present.
In terms of classifier assessment on the mock community, there is no change:
$ head -n 2 summary/ctrl_denoise.ITS2.assess.1s5g.tsv
<SEE TABLE BELOW>
As a table:
#Species |
TP |
FP |
FN |
TN |
sensitivity |
specificity |
precision |
F1 |
Hamming-loss |
Ad-hoc-loss |
|---|---|---|---|---|---|---|---|---|---|---|
OVERALL |
102 |
11 |
1 |
186 |
0.99 |
0.94 |
0.90 |
0.94 |
0.0400 |
0.105 |
Looking at the reports, the read counts are of course different, but also some of the reads assigned a genus-only classification have been removed via the read-correction, so the taxonomy output does not directly match up either.